
- derived from "S"
- statistics software
- a programming language
- extensible
- developed constantly
- platform independent
- well documented
- (almost) essential to know
- free to download

- intuitively usable
- commercially supported
Expressions
- Simple expressions in R are numbers or (character) strings.
- Strings have to be put into double or single quotes.
- Numbers can be used for calculations, strings for string operations.
- Calculating with strings will result in error messages, quite obviously.
Expressions
- Numbers:
3 3.14 - Strings:
"Hallo!" 'Text.' - Calculations:
1+1 2*2 1/3 2-1 10^3
Comparisons
- Comparisons consist of two expressions or objects and a comparison operator.
- Comparisons yield a boolean expression: TRUE or FALSE.
Comparison operators:
- greater than: >
- greater or equal: >=
- less than: <
- less or equal: <=
- unequal: !=
- equal: ==
Comparisons
- Numbers:
3>2 2<4 1<=1 2>=1 3==1 - Strings:
"Hallo!" != 'Text.' "X" == "U" - Boolean expressions:
TRUE == TRUE TRUE != FALSE
Logical Operations
- Logical Operations link two expressions using a logical operator.
- Logical Operations yield a boolean expression: TRUE or FALSE.
- Applicable to: Boolean expressions, numbers
- Not applicable to: strings
Operatoren sind:
- And: & or &&
- Or: | or ||
- Not: prefixed !
- Exklusive or: Function xor(a, b)
Logical Operations
- And:
TRUE & TRUE TRUE && FALSE - Or:
TRUE | TRUE TRUE || FALSE - Not:
! TRUE - Exklusive or:
xor(TRUE, FALSE)
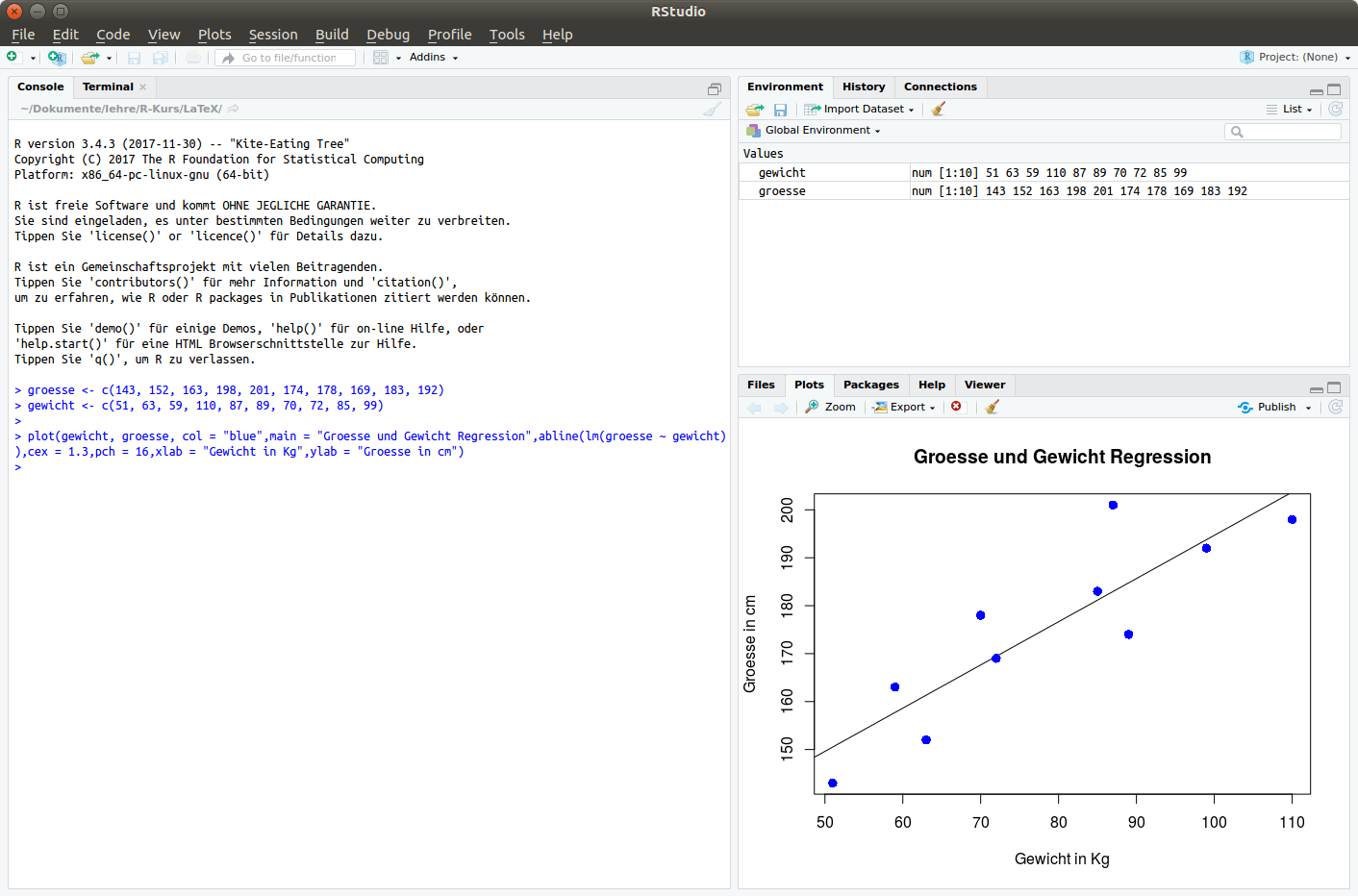
Test it!
- R is available here: https://www.r-project.org/
- R-Studio here: https://www.rstudio.com/
- Both should be already installed on the course notebooks.
Test it!
Variables
- Like any other programming language R knows variables.
- Variables are named storage boxes for data.
- Variable content can be changed.
- Variable types in R are dynamic, i.e. the data type that a variable can contain does not have to be declared beforehand.
- The assignment operator in R is: <- or = oder ->
Variables
Characters that can be used in variable names are alphanumeric characters, numbers, dot and underscore, so:
| variable_number1. | ok |
| .variable_number | ok |
| variable.number | ok |
but
| .1variable_number | not ok, no numbers after "." |
| 1variable_number | not ok, name may not start with a number |
| _variable_number | not ok, name may not start with an underscore |
Assigning to Variables
> varname.1 = 1
> varname.1
[1] 1
> varname.2 <- 1
> varname.2
[1] 1
> "Kurs" -> varname.3
> varname.3
[1] "Kurs"
Overview and Cleaning Up
Listing all variables
# Function ls() will list all defined variables:
> ls()
[1] "alpha" "beta" "df0" "df1" "dg" "g" "x" "m" "M" "pk" "y"
Cleaning up…
# The function rm() will delete variables
> rm(x)
> ls()
[1] "alpha" "beta" "df0" "df1" "dg" "g" "m" "M" "pk" "y"
# Delete everything:
> rm(list=ls())
Output
You can output to screen or a file with the functions print() or cat().
Use cat() to concatenate several output data.
varname1<-'test1'
varname2<-'test2'
print(varname1)
print(varname2)
cat(varname1, varname2)
Heads or Tails
head() and tail() facilitate printing of the first or last lines of an R object.
> seq(1,100)
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
[19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36
[37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54
[55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
[73] 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90
[91] 91 92 93 94 95 96 97 98 99 100
> head(seq(1,100),2)
[1] 1 2
Data Types
| Description | Example | Data type |
|---|---|---|
| Char strings | "R Course" | character |
| integer and real numbers | 1000 | numeric |
| integer numbers | 1L, 2L | integer |
| complex numbers | 6.023x10exp23 | complex |
| logical numbers | TRUE | logical |
| empty | NULL | NULL |
Data Types
> variable1 <- "R Kurs"
# Use the function class() to find out the type
> class(variable1)
> cat("The variable is of type ", class(variable1), ".\n")
The variable is of type character
> variable1 <- 1000L
> cat("Now it is of type ", class(variable1), ".\n")
Now it is of type integer.
Type Casting
The process of converting data types into other data types is referred to as "type casting":
# The function as.integer() converts
# variables to type integer:
> variable2 <- as.integer(1000)
> class(variable2)
[1] "integer"
Overwriting
Variables aren't constants.
# Variables can be overwritten
> variable1 <- "Test"
> variable1 <- 10.7
> cat("The variable is of type ", class(variable1), ".\n")
The variable is of type numeric.
Types
Intern werden Daten von R in expliziteren Typen gespeichert:Internally, R stores data in more verbose types.
var1 <- 10.7
var2<-10
var3 <- as.integer(var1)
var4 <- as.integer(var2)
# In internal memory "numeric" is the same as "double".
# Use typeof() to get the internal mode.
> typeof(var1)
[1] "double"
Which internal and external data types do the used variables have?
Deleting Variables
# Variable can be deleted with rm()
> var1 <- 1000
> rm(var1)
> var1
Fehler: Objekt 'var1' nicht gefunden
Functions
There are many functions that are implemented either in R base or in one of the numerous statistical (and other) packages.
Functions can be used for e.g. statistical calculations, input and output or visualization of data.
User defined functions will be presented later.
Functions
Functions can be used for predefined tasks. Many functions are already implemented in base R, e.g. sum(), mean(), sqrt()
# Calculate sums with sum()
> sum(1000, 1)
[1] 1001
# Calculate the arithmetic mean with mean()
> mean(c(1, 2, 3, 4, 5)) # this is a vector...
[1] 3
Square Root
# Calculate the square root
> sqrt(49)
[1] 7
Logarithms
# Common logarithm functions
# Logarithm base 10
> log10(1000)
[1] 3
# Logarithm base 2
> log2(64)
[1] 6
# natural logarithm
> log(100)
[1] 4.60517
# Exponent
> exp(4.60517)
[1] 99.99998
# NB.: rounding error!
Help!
R does also contain a user manual: ?sqrt or help(sqrt) will show the manual page for the function sqrt().
# Was does this do?
help(help)
'q' will exit the (text based) help display.
Data Structures
In R data is organized in data structures, e.g. vectors, lists, matrices…
- Vector
- Matrix
- Array
- Data frame
- List
- Factor
Vectors
- Vectors are elementary parts of R
- Vectors consist of one or more ordered elements
- Vectors can hold all data types, e.g. numeric, character, logical
- All elements within a given vector have to be of the same type!
Vectors
- In R almost everything is implemented based on vectors.
- Vectors can be any data type. But again: Vectors can't hold elements of mixed types!
- The most simple vectors in R consist of only one element.
Vectors
# You create a character type vector like this:
> print("Kurs") # e.g.
[1] "Kurs"
# It can also be assigned to a variable:
> var1 <- "Kurs"
[1] "Kurs"
# Check if var1 is a vector (or anything else) by preceding "is.":
> is.vector(var1)
[1] TRUE
Vectors
# Casting a vector to a certain type can be done with "as."
> var2 <- pi
> var3 <- as.integer(var2)
> var3
[1] 3 # Information is lost here!
> class(var3)
[1] "integer"
Vectors
Numeric vectors with several elements:
# More than one way to create a numeric vector
> x <- c(1, 2, 3, 4, 5)
> x
[1] 1 2 3 4 5
> x <-seq(from = 1, to = 5, by = 1))
> x
[1] 1 2 3 4 5
> x <- c(1:5)
> x
[1] 1 2 3 4 5
Vectors
The easiest way to create a vector is the c() function (for combine or concatenate).
# numerischer Vector
> x <- c(1:100)
# "character" Vector
> x <- c("Ich", "brauche", "eine", "Pause")
> x
[1] "Ich" "brauche" "eine" "Pause"
# Boolean Vector
> x <- c(FALSE, TRUE, T, F)
> x
[1] FALSE TRUE TRUE FALSE
Vectors
Mixed vectors:
> x <- c(1, 2, TRUE, "Kaffee")
> x
# All elements have to be of the same type.
# Thus "character" type is enforced in this case.
>is.character(x)
[1] TRUE
Vectors
# Combining vectors
> x <- c(1:5)
> y <- c(x, 6:10)
> y
[1] 1 2 3 4 5 6 7 8 9 10
> length(y)
[1] 10
Vectors
To repeat a certain object use the function rep().
> x <- rep(3, 12)
> x
[1] 3 3 3 3 3 3 3 3 3 3 3 3
> length(y)
[1] 12
Calculations with Vectors
Calculating with vectors happens element by element:
> x <- c(1:5)
> y <- x*2
> y
[1] 2 4 6 8 10
> z <- x + x
> z
[1] 2 4 6 8 10
> z <- x ^2
> z
[1] 1 4 9 16 25
> sum(x) # an aggregating function!
[1] 15
> mean(x)
[1] 3
Indexing Vectors
Syntax: Object[Index-Vector]
> x <- c(1:5)
> x[3] # index third vector element
[1] 3
> x[c(4, 1)] # index elements four and one (order is important!).
[1] 4 1
> x[-c(4, 1)] # index everything BUT elements four and one
[1] 2 3 5
Other Data Structures
In R all data structures belong to certain classes.
Vectors are most important. Everything else can be seen as combinations of vectors into a certain structure.
Matrices
Data in a matrix is organized in two-dimensional form.
All elements have to be of the same type, still.
Numeric elements are the most common type used in matrices.
Datasets – Data Frames
Dataframes are similar to matrices but can hold different data types in different columns
Lists
Lists are R objects that can hold several elements of arbitrary types, including vectors, numbers, other lists…
Factors
Factors are vector-like R objects that hold categorized data. The levels or categories valid for the factor are defined at its creation.
If the factor is created for levels "a" and "b" you can add another "a" but you can't add a "c".
Useful for property-like data with several set options only, e.g. "male/female", "wildtype/mutant" or "A/G/T/C".
Arrays
Arrays are R objects that can hold data organized in more than two dimensions.
An array with the dimensions (2, 3, 4) can e.g. be interpreted as a stack of four matrices with two rows and three columns each.
Will not be discussed here.
Datentypen
| Typ | Dim | Mix | Named |
|---|---|---|---|
| Vector | 1 | FALSE | FALSE |
| Matrix | 2 | FALSE | TRUE |
| Data frame | 2 | TRUE (columnwise) | TRUE |
| List | 1 (but...) | TRUE | FALSE |
| Factor | 1 | TRUE | FALSE |
| Array | n | FALSE | TRUE |
Creating a Matrix
Matrices are created with the function matrix().
Syntax: matrix(data, nrow, ncol, byrow, dimnames)
- data Input-Vector, will supply the matrix elements.
- nrow Number of rows
- ncol Number of columns
- byrow If TRUE the matrix will be built row by row instead of by column.
- dimnames List of vectors containig row and column names
Creating a Matrix
# elements filled in by column
> x <- matrix(1:10, nrow = 2, ncol = 5)
> x
[,1] [,2] [,3] [,4] [,5]
[1,] 1 3 5 7 9
[2,] 2 4 6 8 10
# elements filled in by row
> x <- matrix(1:10, nrow = 2, ncol = 5, byrow = TRUE)
> x
[,1] [,2] [,3] [,4] [,5]
[1,] 1 2 3 4 5
[2,] 6 7 8 9 10
Creating a Matrix
# naming columns and rows
> rownames <- c("Zeile1", "Zeile2")
> colnames <- c("Spalte1", "Spalte2", "Spalte3", "Spalte4", "Spalte5")
> y <- matrix(1:10, nrow = 2, ncol = 5,
byrow = TRUE, dimnames = list(rownames, colnames))
> y
Spalte1 Spalte2 Spalte3 Spalte4 Spalte5
Zeile1 1 2 3 4 5
Zeile2 6 7 8 9 10
Indexing Matrices
Matrix elements can be addressed by row- and column index.
# Index element in col 3, row 2
> y[2,3]
> [1] 8
To index whole columns or rows, leave out the other dimension index.
# Index column 4
> y[,4]
Zeile1 Zeile2
4 9
Calculations with Matrices
This can be used for calculations, of course.
# Hier wird der Mittelwert aller Elemente in Zeile1 in Spalte1 bis Spalte3 berechnet
# Calculate the average of elements in columns 1 to 3, row 1
> mean(y[1,c(1, 2, 3)])
# or
> mean(y[1,1:3])
Non-aggregating calculations are also done per element.
> y*2
Column1 Column2 Column3 Column4 Column5
Row1 2 4 6 8 10
Row2 12 14 16 18 20
Dataframes
Dataframes are similar to matrices but –in contrast– can hold different data types
This is not possible in matrices, as the following example will demontrate.
# In matrices all elements are born equal ;)
> x <- matrix(c(1:5, c("a", "b", "c", "d", "e")), 5, 2)
> x
[,1] [,2]
[1,] "1" "a"
[2,] "2" "b"
[3,] "3" "c"
[4,] "4" "d"
[5,] "5" "e"
> y <- x[2,1]
> class(y)
[1] "character"
# The numbers are demoted to chars, here
Mixed Dataframes
In a dataframe elements of different type can be combined. Column names (colnames) shouldn't be empty row names should be unique or empty.
# Same data as before:
> d <- data.frame(ersteSpalte = 1:5, zweiteSpalte = c("a", "b", "c", "d", "e"))
ersteSpalte zweiteSpalte
1 1 a
2 2 b
3 3 c
4 4 d
5 5 e
But: elements sharing a column do have to be of the same type!
Dataframe Example
In this example we consider 5 plants of a mutant Arabidopsis line. We need to store plant number and number of well developed seeds.
> dfrm <- data.frame(Plant_No =
c("At1", "At2", "At3", "At4", "At5"),
fertile_seeds = c(122, 345, 215, 90, 77),
stringsAsFactors = FALSE)
Pflanzen_Nr fertile_Samen
1 At1 122
2 At2 345
3 At3 215
4 At4 90
5 At5 77
Dataframe Structure
With the function str() we can check a dataframe's structure:
> str(dfrm)
'data.frame': 5 obs. of 2 variables:
$ Pflanzen_Nr : chr "At1" "At2" "At3" "At4" ...
$ fertile_Samen: num 122 345 215 90 77
str() functioniert übrigens nicht nur mit Dataframes.
Dataframe Summary
The Function summary() will print out a statistical summary of a dataframe.
> summary(drfm)
Pflanzen_Nr fertile_Samen
Length:5 Min. : 77.0
Class :character 1st Qu.: 90.0
Mode :character Median :122.0
Mean :169.8
3rd Qu.:215.0
Max. :345.0
Dataframe Column Extension
We can add data to a dataframe. Here we will include a column for flowering time in days.
# The $ addresses a column by name
# It's added since it's not there yet.
> data$Bluehzeit_Tage <- c(5, 7, 6, 10, 8)
> data <- data
> data
Pflanzen_Nr fertile_Samen Bluehzeit_Tage
1 At1 122 5
2 At2 345 7
3 At3 215 6
4 At4 90 10
5 At5 77 8
Dataframe Row Extension
To add new rows to a dataframe use the function rbind(). It combines two dataframes of the same layout.
# First we need a new dataframe of the same format with data to add
> newdata <- data.frame(Pflanzen_Nr = c("At6", "At7", "At8"),
fertile_Samen = c(99, 45, 102), Bluehzeit_Tage = c (7, 11, 13),
stringsAsFactors = FALSE)
# Then we can connect both frames
# rbind == row bind
> data_final <- rbind(data, newdata)
> print(data_final)
Plant_No fertile_seeds Bluehzeit_Tage
1 At1 122 5
2 At2 345 7
3 At3 215 6
4 At4 90 10
5 At5 77 8
6 At6 99 7
7 At7 45 11
8 At8 102 13
Indexing Dataframes
Quite similar to indexing matrices.
# Extracting the first two rows
> print(data_final[c(1, 2),]) # or [1:2,]
Plant_No fertile_seeds Bluehzeit_Tage
1 At1 122 5
2 At2 345 7
# Here we extract flowering time of plants 5 and 8
> print(data_final[c(5, 8), c(1, 3)])
Plant_No Bluehzeit_Tage
5 At5 8
8 At8 13
# addressing columns by name:
> data_final$fertile_seeds[2]
[1] 345
Lists
Lists are created with the list() function. They can be seen as a collection object for different components, possibly belonging to different data types.
# Defining a list holding a vector and a matrix.
# E.g. expression values of 3 genes in two replicates.
> newList <- list(c("WUSCHEL", "CLAVATA3", "TOPLESS"),
expr = matrix(c(101, 59, 120, 289, 310, 278, 789, 1020, 812),
nrow = 3, ncol = 3, byrow = TRUE))
> neueListe
[[1]]
[1] "WUSCHEL" "CLAVATA3" "TOPLESS"
$expr
[,1] [,2] [,3]
[1,] 101 59 120
[2,] 289 310 278
[3,] 789 1020 812
Lists can contain other lists as well!
Factors
Factors assign data elements to distinct categories or levels which can be useful in statistical analyses.
E.g. you can define a factor that can only hold the values "male" and "female" or 1 and 2 or "wiltype" and mutant. Internally the levels are stored as integer values.
Factors are created with the function factor().
Factors
# Create a vector with the data
> genotype <- c("wildtype", "wildtype", "mutant", "wildtype", "mutant")
# That will become a two level factor:
> g <- factor(genotype)
[1] wildtype wildtype mutant wildtype mutant
Levels: mutant wildtype
# You now cannot add a hedgehog to it anymore:
> g[6]<-'hedgehog'
Warning:
In `[<-.factor`(`*tmp*`, 6, value = "hedgehog") :
invalid factor level, NA generated
> g
[1] wildtype wildtype mutant wildtype mutant NA
Levels: mutant wildtype
Flow Control and Loops
Control constructs and loops are important parts of all programming languages. They control program flow by conditional execution of code blocks and repetition.
# If a condition is TRUE expression1 is evaluated, on false expression2 is used.
# The other expression respectively is skipped entirely.
> if(condition) {expression1} else {expression2} # (1)
# A more R-ish way to put it:
> ifelse(condition, expression1, expression2) # (2)
Conditional Execution
An example:
> TheTime <- "10 h"
> Breakfast <- "9 h"
> Lunch <- "12 h"
> if(TheTime == Lunch || TheTime == Breakfast) {print("Pause")}
else{print("Go on working")}
[1] "Go on working"
An example for ifelse()
ifelse() is a function, accepting a conditional expression and two alternative code expressions. It is vectorized, i.e. can work on a vector element by element.
.
> x <- c(1:10)
> ifelse(x > 5, ">5", "<=5")
[1] "<=5" "<=5" "<=5" "<=5" "<=5" ">5" ">5" ">5" ">5" ">5"
Loops
Loops can carry out functions or code blocks repeatedly. There are several possibilities for loops in R:
| repeat{expression} | Repeat expression until stopped |
| while(condition){expression} | Repeat expression while condition is TRUE |
| for(i in M){expression} | Repeat expressions for every element i in assemblage M |
| next | Skip to next iteration |
| break | exit loop immediately |
Repeat Example
# Increment i by 1 until 10 is reached
> i <- 0
> repeat{
+ i <- i+1
+ print(i)
+ if(i == 10) break
+ }
[1] 1
[1] 2
[1] 3
[1] 4
[1] 5
[1] 6
[1] 7
[1] 8
[1] 9
[1] 10
While Example
# Usage example for while
# (You can skip the braces for single expressions.)
> i <- 0
> while(i < 10)
+ i <- i +1
> i
[1] 10
For Example
> i <- 0
> x <- c(1:10)
> for(i in x)
+ print(i + 1)
[1] 2
[1] 3
[1] 4
[1] 5
[1] 6
[1] 7
[1] 8
[1] 9
[1] 10
[1] 11
Input and Output of Files
R can access data outside the R system as well as export data to files.
File I/O
It is important to define and be aware of the currently used folder. To set a new working directory you can use the function setwd(). To find out where in the file system you currently are, use getwd()
To see what is in a folder use dir().
A "wd" might be C:/Users/You/R/subfolder on Windows or /home/You/R/subfolder on Unix-likes.
The easiest way in R to read data from a file is read.table(). The function is suitable for reading in a data frame stored as a file in tabular form.
File I/O
Important options of read.table()- file – Dateiname und Pfad (letzerer nur, wenn nicht im working directory vorhanden)
- header – deklariert, ob Spaltennamen vorhanden sind, die default Einstellung ist FALSE
- sep – definiert die Trennzeichen zwischen den Spalten. Default ist ein Leerzeichen oder Tab (" ", "\t")
- dec – definiert, welches Trennzeichen Dezimalstellen trennt, die Voreinstellung ist "."
- Informationen finden sich in der Hilfe ?read.table
Reading in Data
> ?read.table()
read.table(file, header = FALSE, sep = "", quote = "\"'",
dec = ".", numerals = c("allow.loss", "warn.loss", "no.loss"),
row.names, col.names, as.is = !stringsAsFactors,
na.strings = "NA", colClasses = NA, nrows = -1,
skip = 0, check.names = TRUE, fill = !blank.lines.skip,
strip.white = FALSE, blank.lines.skip = TRUE,
comment.char = "#",
allowEscapes = FALSE, flush = FALSE,
stringsAsFactors = default.stringsAsFactors(),
fileEncoding = "", encoding = "unknown", text, skipNul = FALSE)
Writing out Data
To output data write.table() is used.
write.table(x, file = "", append = FALSE, quote = TRUE, sep = " ",
eol = "\n", na = "NA", dec = ".", row.names = TRUE,
col.names = TRUE, qmethod = c("escape", "double"),
fileEncoding = "")
?write.table
Data Exchange with MS Excel
To exchange data between Excel (et al.) and R CSV files are a valid solution. The appropriate functions in R are:
read.csv(file, header = TRUE, sep = ",", quote = "\"",
dec = ".", fill = TRUE, comment.char = "", ...)
read.csv2(file, header = TRUE, sep = ";", quote = "\"",
dec = ",", fill = TRUE, comment.char = "", ...)
read.delim(file, header = TRUE, sep = "\t", quote = "\"",
dec = ".", fill = TRUE, comment.char = "", ...)
read.delim2(file, header = TRUE, sep = "\t", quote = "\"",
dec = ",", fill = TRUE, comment.char = "", ...)
write.csv(...)
write.csv2(...)
Accordingly use write.csv() for output.
Exercise Data in R
As an example, access the famous "iris" dataset.
?iris
iris {datasets}R Documentation
Edgar Anderson's Iris Data
Description
This famous (Fisher's or Anderson's) iris data set gives the measurements in
...
> head(iris)
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
Sepal.L.. Sepal.W.. Petal.L.. Petal.W.. Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
> write.table(iris, file = "iris_data.txt", sep = " ")
Iris
You can read the same dataset with read.table(), now:
> test <- read.table("iris_data.txt", header = TRUE, sep = " ")
> head(test)
Sepal.L.. Sepal.W.. Petal.L.. Petal.W.. Species
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3.0 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
R Packages
- R is extensible.
- There are useful functions for a lot of applications, organized in packages.
- To make use of them you have to load the respective package first.
- To be able to load a package it has to be installed. Installation can be done system-wide (admin's job) or into a presonal library.
Installing R packages
Before you can use functions from a package it might have to be installed first.
# To install a package this will be sufficient:
> install.packages('paketname')
# To use a certain folder explicitely:
> install.packages('paketname', lib='/home/username/myRlib')
CRAN-Mirrors
- Packages are installed from a CRAN mirror server on the internet, per default.
- CRAN: "Comprehensive R archive network"
- CRAN is the central place to look for published R packages. To keep server load low there are several, globally distributed mirror servers sharing the same content.
- There are other package repositories like BioConductor or Github…
Installing R Packages
An Example:
> install.packages('ggplot2')
Installiere Paket nach '/home/username/R/x86_64-pc-linux-gnu-library/3.4'
(da 'lib' nicht spezifiziert)
– Bitte einen CRAN Spiegel fuer diese Sitzung auswaehlen –
versuche URL 'https://ftp.gwdg.de/pub/misc/cran/src/contrib/ggplot2_2.2.1.tar.gz'
Content type 'application/octet-stream' length 2213308 bytes (2.1 MB)
==================================================
downloaded 2.1 MB
* installing *source* package 'ggplot2' ...
** Paket 'ggplot2' erfolgreich entpackt und MD5 Summen ueberprueft
...
* DONE (ggplot2)
Die heruntergeladenen Quellpakete sind in
'/tmp/RtmpCnOyO7/downloaded_packages'
Loading R-Packages
Once installed a package has to be loaded into the workspace:
> library(paketname) # no quotes
> library(help=ggplot2) # an overview of available functions in the package
Exercises
Exercises on creation, access and calculation with vectors
Exercise 1:
Create a vector containing the numbers 3, 9, 7, 6, 4, 1.
Then calculate the mean of the elements in the vector and the square root of the different elements.
Exercise 2:
Create a vector containig the numbers 1, 10 and 100 with data type of integer.
Use two different methods. Test the data type by using an R function call.
Exercise 3:
Create a vector with the elements "Easy", "work", "with", "R".
What's its data type?
Exercise 4:
Create a vector that contains the odd numbers between 1 and 100.
Try to find the simplest way to do so.
Exercise 5:
Create a vector x containig the numbers from 1 to 5 and a vector y with the numbers 7 to 20.
Combine both vectors to get vector z. Delete x and y from memory.
Sort z in decreasing order.
Exercise 6:
Calculate the mean of elemnts number 2, 7 and 9 of vector z (s. Ex. 5).
Exercises on Matrices
Exercise 1a:
Create this matrix as M1:
[,1] [,2]
[1,] 1 6
[2,] 2 7
[3,] 3 8
[4,] 4 9
[5,] 5 10
Exercise 1b:
Do the same under the name of M2, but row-wise.
Exercise 1c:
Get the element in column 2, row 3 from that matrix (s. 1b, M2). (Should be number 6.)
Exercise 1d:
Subtract 2 from every element of the second column. Calculate the mean of that new vector.
Exercise 1e:
Create a matrix M3 from M2 where the numbers in column 2 are replace by the numbers in column 1, each subtracted by 2.
Exercise 1f:
Add a sixth row to M3 containing the elements 12 and 14. And another row with 20 and 25.
Exercise 2:
Create a matrix with two rows and two columns containing numbers 1 to 4, row-wise.
Row names should be "a1" and "a2", column names "b1" and "b2".
Exercise 3:
Create the following matrix using the shortest way you can think of.
[,1] [,2] [,3] [,4] [,5]
[1,] 100 80 60 40 20
[2,] 98 78 58 38 18
[3,] 96 76 56 36 16
[4,] 94 74 54 34 14
[5,] 92 72 52 32 12
[6,] 90 70 50 30 10
[7,] 88 68 48 28 8
[8,] 86 66 46 26 6
[9,] 84 64 44 24 4
[10,] 82 62 42 22 2
Exercises on Dataframes
Exercise 3a:
Create the following data frame as "data".
Plant_No fertile_Seeds aborted_Seeds
1 At1 377 10
2 At2 879 30
3 At3 216 41
4 At4 93 71
5 At5 98 22
6 At6 103 6
Exercise 3b:
Print out the statistical summary for "data".
Exercise 3c:
We forgot some data. Add the column "Counted_silics".
Use 6, 18, 4, 3, 2 and 2 silics accordingly.
Exercise 3d:
Sort the data by increasing number of counted silics. Use the function order().
Exercise 4:
Open the iris dataset as dataframe "d" for the following exercises.
Exercise 4a:
View the top of the dataset with head().
Now let's access all rows holding data for the species "virginica". One possibility is to use the function which(). which() will generate a numeric vector whose elements correspond to TRUE in a corresponding logical vector.
First create that numeric vector and a new dataframe "subset" in a second step.
Exercise 4b:
Create a new dataframe "subset2" from "subset" that contains all virginica species with a Petal width below 2.
Exercises with Loops
Exercise 1
Use the function ifelse() to print out "Nope" if an element of a vector containig the numbers from 10 to 20 is not 16 and "Hooray" if it is.
Exercise 2
Use repeat() and break to implement a count-down from 9 to 0.
Exercise 3
Create a loop that multiplies each of the numbers from 1 to 10 with 3 and print them out.
Exercises with other functions
Exercise 1
Use the function grep() to get alle elements from the following vector containing "RKURS":
x <- c("KURSFREITAGHEUTE", "HEUTERKURSTOLL", "RKUSISTEIN",
"ALLERKURSESINDAUSGEBUCHT", "ALLESKURSIVODER")
Exercise 2
Please generate the following matrix:
> x <- matrix(c(1:100), nrow = 10).
Use the function apply() to read out the smallest number of each column..